BREAKING NEWS: A revolutionary 3D deep learning model has been unveiled, transforming the way scientists analyze fruit tissue microstructure. Published on 5 July 2025, this groundbreaking research from KU Leuven, led by Pieter Verboven, sets a new standard in plant biology, achieving an unprecedented accuracy in tissue segmentation that far surpasses previous methods.
This innovative approach employs X-ray micro-CT imaging, which allows for non-destructive 3D visualization of plant tissues. Traditional methods have struggled with extensive prep time and limited field views, making this development crucial for advancing research into vital metabolic processes in plants.
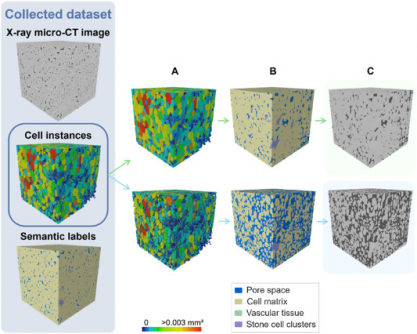
The newly developed model utilizes a 3D panoptic segmentation framework based on the 3D extension of Cellpose and a 3D Residual U-Net, successfully automating the labeling and quantification of fruit tissue architecture. This advancement facilitates faster and more precise studies of plant physiology and storage behavior, with significant implications for agriculture and food science.
In rigorous evaluations, the model achieved a remarkable Aggregated Jaccard Index (AJI) of 0.889 for apples and 0.773 for pears, outperforming the previous 2D methods, which scored 0.861 and 0.732, respectively. The model excelled in segmenting pore spaces and cell matrices, while effectively identifying vascular structures and stone cell clusters.
Visual validations confirmed the model’s accuracy, showing detailed segmentation of vascular bundles in popular apple varieties like Kizuri and Braeburn, and realistic representations of stone cell clusters in Celina and Fred pears. With a Dice Similarity Coefficient (DSC) reaching up to 0.90, this model proves to be the most complete and automated method for quantifying plant tissue microstructure to date.
The implications of this advanced technology extend beyond fruit analysis. It offers a powerful, non-destructive tool for studying how microscopic structures influence water, gas, and nutrient transport in various crops. This can significantly reduce manual labor in research, enhancing accuracy in tissue characterization.
As global agriculture faces challenges such as climate change and food security, this technology stands out as a scalable solution for studying tissue development and stress responses across diverse plant species. Its compatibility with standard X-ray micro-CT instruments makes it an accessible option for researchers worldwide, enabling the integration of artificial intelligence into plant anatomy studies.
WHAT’S NEXT: Researchers and plant scientists are encouraged to adopt this cutting-edge model, which not only streamlines plant tissue analysis but also opens doors to new discoveries in plant physiology. As the agricultural sector seeks innovative solutions to improve crop resilience and quality, developments like these will be critical in shaping the future of food science.
This breakthrough research is featured in the esteemed journal Plant Phenomics, which is committed to advancing plant research and connecting diverse scientific domains. With the potential to revolutionize plant phenotyping, this deep learning model could play a pivotal role in the future of agriculture.
Stay tuned for further updates on this exciting development and its implications for the scientific community and beyond.